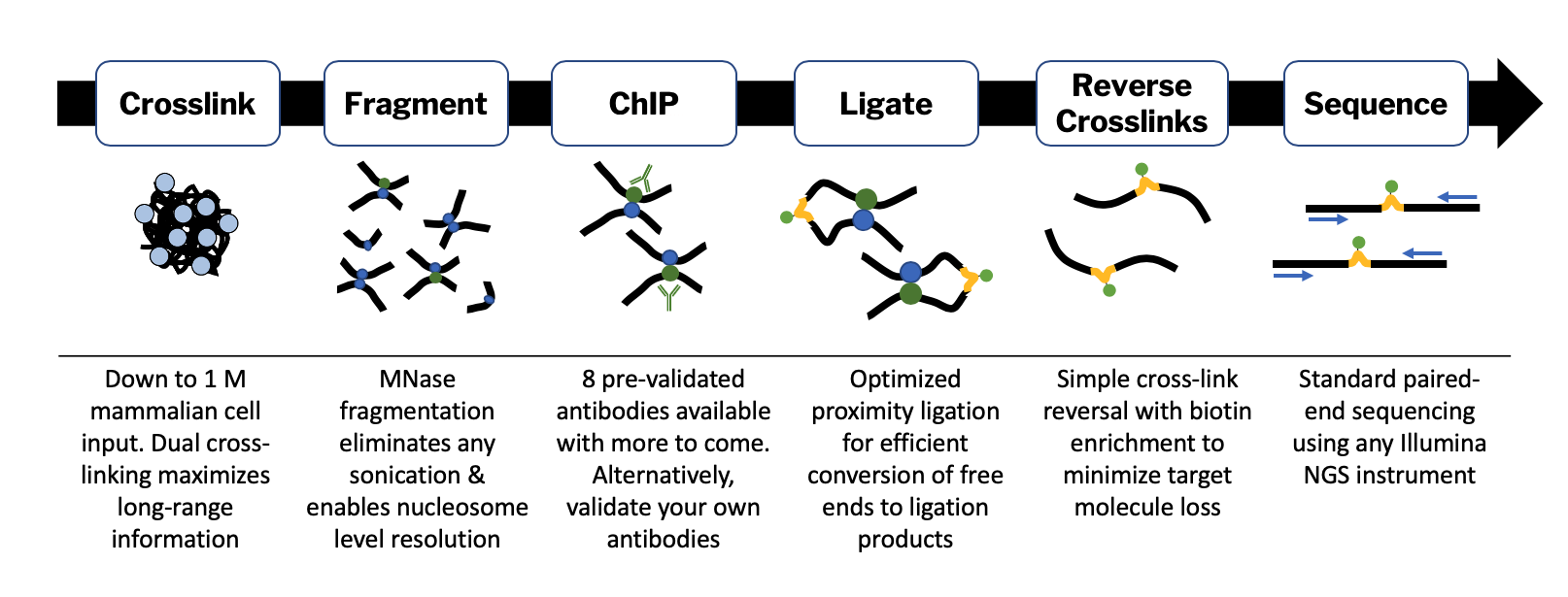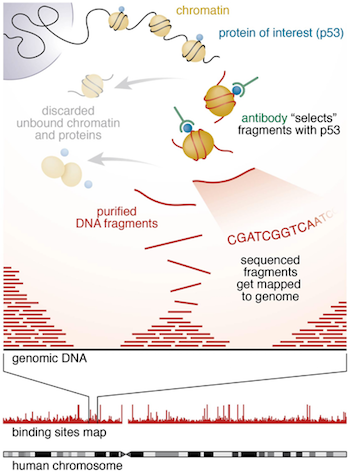
Introduction to ChIP-seq and directory setup | Introduction to ChIP-Seq using high-performance computing

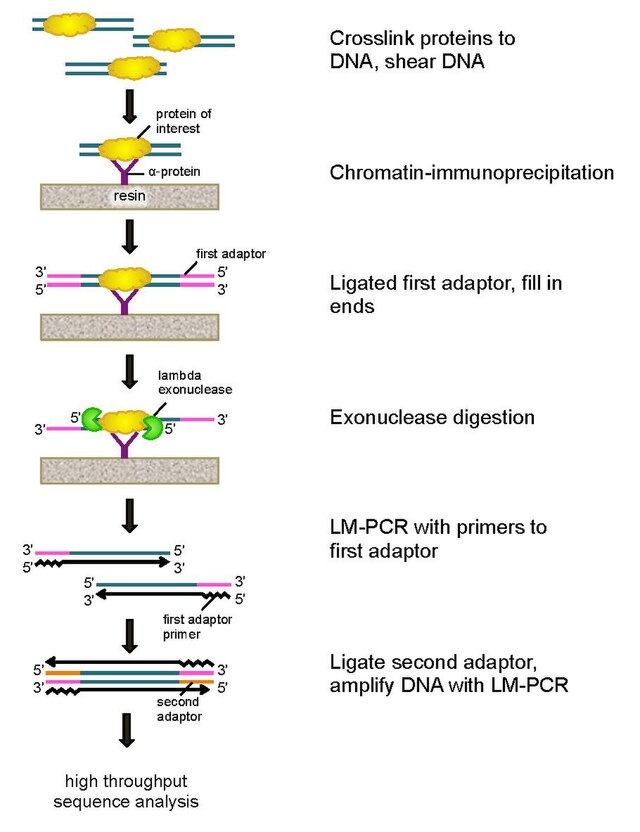
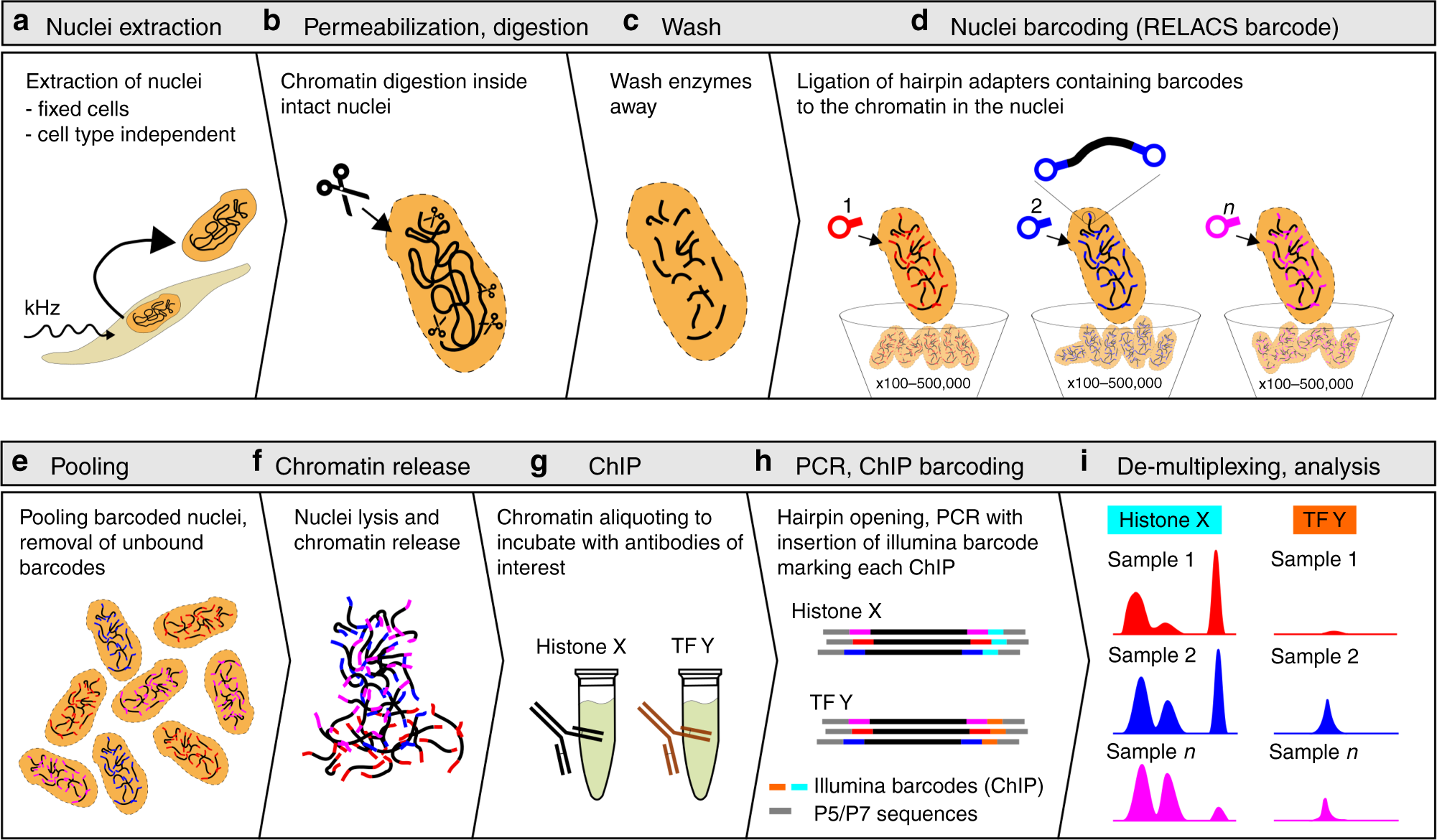
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance
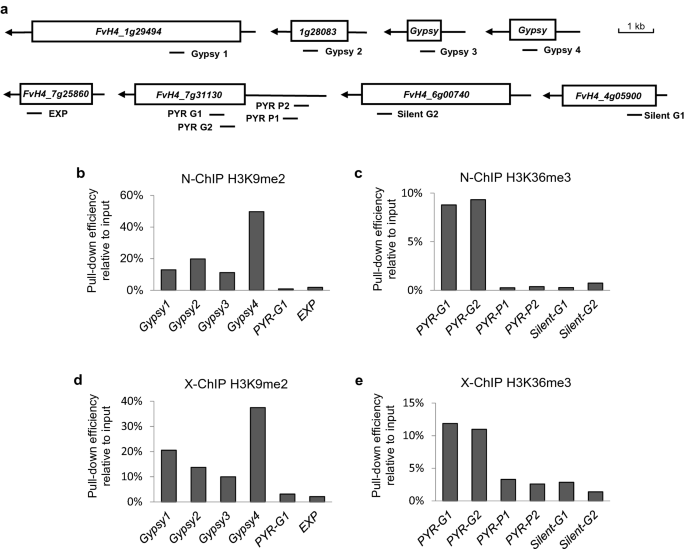
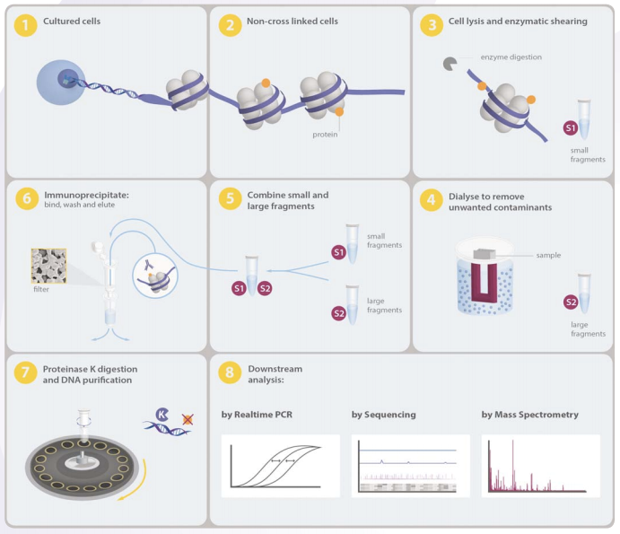
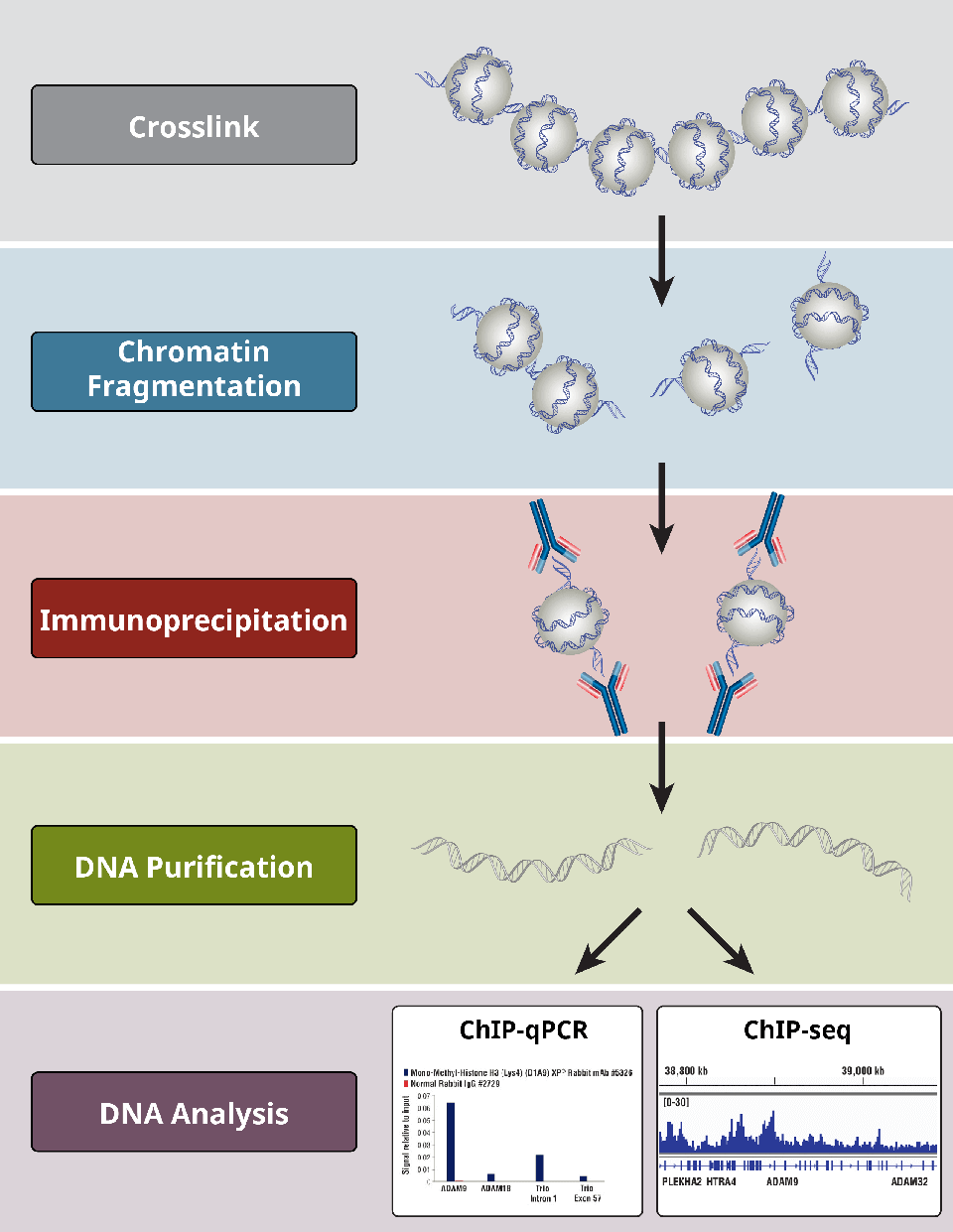
A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits | Plant Methods | Full Text
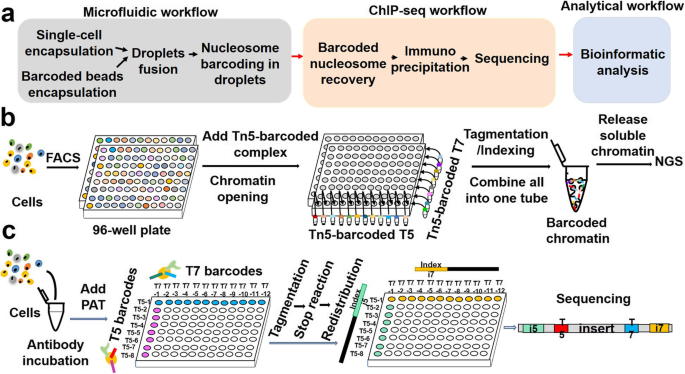
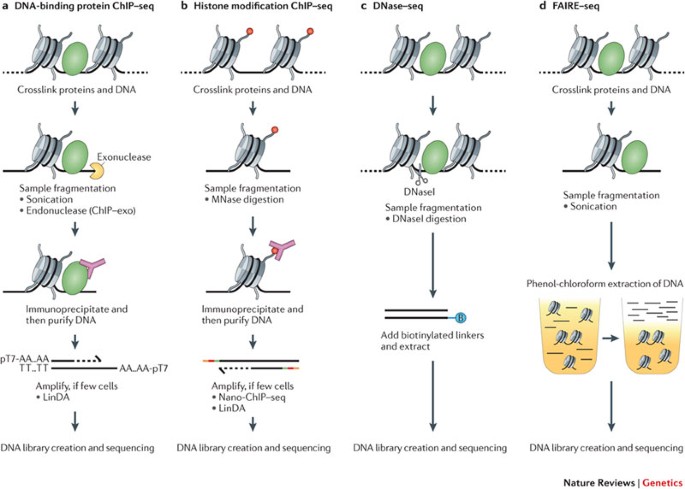
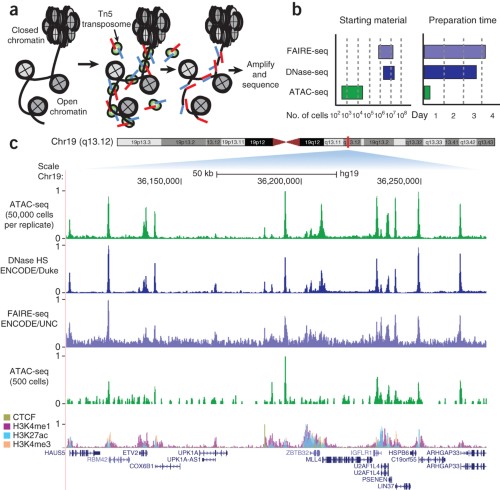
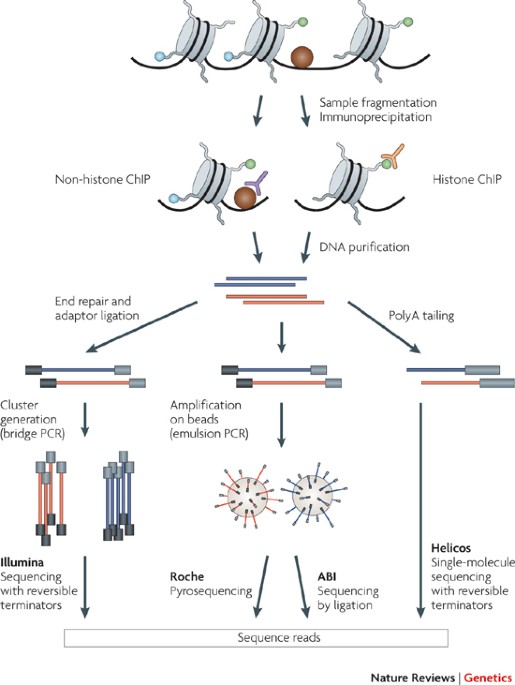
Profiling chromatin regulatory landscape: insights into the development of ChIP-seq and ATAC-seq | SpringerLink
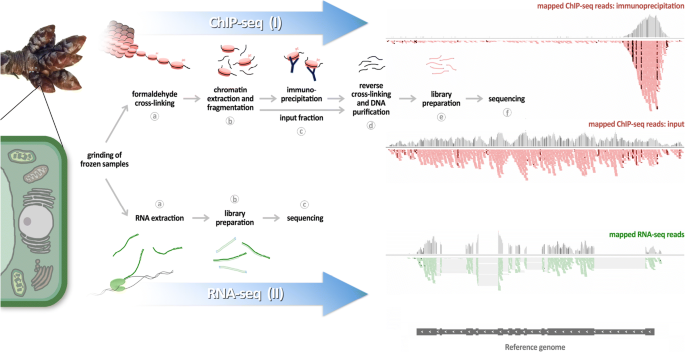
ChIP-seq and RNA-seq for complex and low-abundance tree buds reveal chromatin and expression co-dynamics during sweet cherry bud dormancy | SpringerLink